Theresia Dunzendorfer-Matt 

Research interests 
Structure
and function of proteins in oncogenic pathways
We are interested in the structure and
function of proteins and their functional interactions. The Ras-Regulator
Neurofibromin, which is functionally impaired in the rare disease
Neurofibromatosis Type I (NF1) but also frequently mutated in a growing number
of sporadic tumors is in the center of our interest. Current research includes
analysis of interaction partners and their effect on protein function,
crystallisation of Neurofibromin protein complexes and subsequent x-ray
analysis as well as additonal approaches utilizing nuclear magnetic resonance
(NMR) spectroscopy (collaboration with Martin
Tollinger, University of Innsbruck).
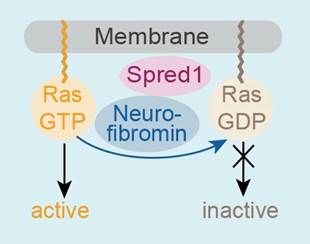 Neurofibromin acts on Ras
at the plasma membrane
Neurofibromin acts on Ras
at the plasma membrane
and is recruited from the cytosol by interaction with Spred1.
This recruitment is required for efficient Ras inactivation.
NF1 patients have mutations in the giant NF-1 gene (encoding the 300 kDa Neurofibromin protein) and share some mild symptoms with the rasopathic disorder Legius Syndrome caused by mutations in the Spred1 gene. The encoded Spred1 protein acts as a recruitment factor and translocates the Ras-specific GAP (GTPase activating protein) Neurofibromin to the plasma membrane. We have identified the Neurofibromin GAP domain as the Spred1 binding site. The Spred1(EVH1) domain specifically recognizes residues located directly N-terminal and C-terminal to the minimal GAP domain forming GAPex which we have assigned a role as a new protein-protein interaction motif (collaboration with Frank McCormick, UCSF). In some patients suffering from NF1, this interaction is disturbed. Mutations detected in Legius Syndrome patients also prevent the formation of this complex. We have analyzed potential effects on enzymatic GAP activity and the stability of this complex in vitro and are aiming at the atomic details of this interaction by a structural approach.
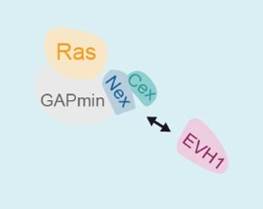 The Spred1(EVH1) domain
recognizes the extra domain
The Spred1(EVH1) domain
recognizes the extra domain
of the Neurofibromin GAP region formed by Nex and Cex.